Participants
US Department of Agriculture, Agriculture Research Service, Peoria, Illinois, USA (Dr. Robert H. Proctor)
Section for Chemistry and Toxicology, Norwegian Veterinary Institute, Oslo, Norway (Dr. Silvio Uhlig)
Background and state of the art
Fumonisins are polyketide-derived mycotoxins produced by some filamentous fungi in the genus Fusarium. The fungus is a widespread pathogen of maize plants, on which it causes ear and stalk rot as well as seedling blight. Fumonisins cause several fatal animal diseases, including leukoencephalomalacia in horses, pulmonary edema in swine, and cancer in rats and mice. In addition, fumonisins are suspected to cause human esophageal cancer and neural tube defects. The B-series fumonisins, fumonisin B1, B2, B3, and B4, are generally the most abundant fumonisins in naturally contaminated maize kernels and in pure cultures of F. proliferatum and F. verticillioides, although about 30 different analogues have been described.
Fumonisin biosynthetic enzymes identified to date are all encoded at one locus, the 17-gene FUM cluster. The FUM cluster also encodes a protein that regulates expression of the cluster genes and proteins involved in transport of fumonisins or biosynthetic precursors across membranes. A biosynthetic pathway for fumonisin production has been proposed based on a combination of biochemical and genetic evidence, including production of fumonisin analogues by mutant strains of Fusarium in which individual FUM genes have been inactivated (Figure 1). In the pathway, a linear, 20-carbon polyketide is formed first. Subsequently, an amino group, up to four hydroxyl functions, and two tricarboxylic acid moieties are added to various positions along the polyketide backbone. Although there is good evidence for most of the steps in the pathway, the products of the first two steps, a 20-carbon linear polyketide and the condensation product of the polyketide and the amino acid alanine, have not yet been identified.
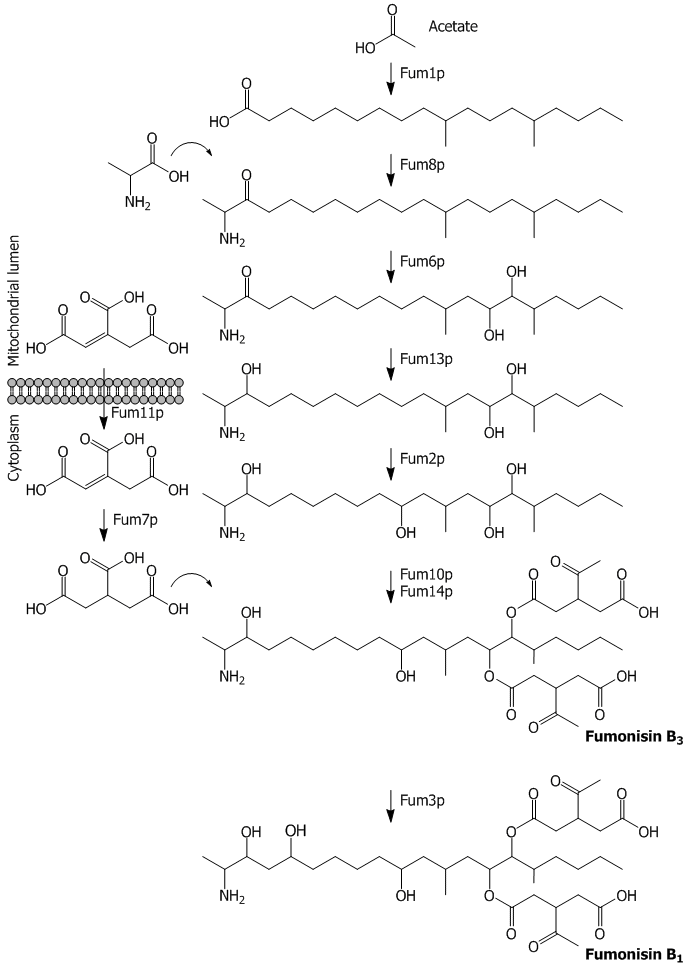
Figure 1. Proposed biosynthetic pathway for fumonisin B1 production showing functions of FUM gene/proteines (reproduced from Alexander et al., 2009, Toxin Reviews 28, 198-215)
Strains of Fusarium in which the FUM cluster genes FUM1, FUM6 and FUM8 have been mutated are the only FUM gene mutants that do not produce any known fumonisin analogs. Thus, the genes most likely encode enzymes that catalyze early steps in fumonisin biosynthesis. FUM1 encodes a polyketide synthase and, therefore, is most likely responsible for synthesis of the linear polyketide that forms the backbone of fumonisins. FUM8 encodes an alpha-oxoamine synthase, a class of enzymes that normally catalyze condensation of an amino acid and an acyl compound (e.g. fatty acid or polyketide). Based on this and the chemical structure of fumonisins, it is likely that the FUM8 enzyme catalyzes the condensation of the linear polyketide and alanine to form a 22-carbon compound (2-amino-12,16-dimethyleicosane-3-one). This proposed compound is structurally highly similar to 2-amino-14,16-dimethyloctadecan-3-ol (2-AOD-3-ol), which is produced by F. avenaceum and was recently identified at NVI. Thus, mutant strains of F. verticillioides in which FUM8 has been inactivated should produce the linear polyketide but not the predicted product of the FUM8 enzyme-catalyzed reaction. The FUM6 gene encodes a cytochrome P450 monooxygenase, a class of enzymes that often catalyze hydroxylation reactions in fungi. Based on this and on the fact that all other hydroxyl groups along to fumonisin molecule result from the activity of other FUM enzymes, it is likely that the FUM6 enzyme catalyzes hydroxylation of fumonisins at carbon atoms 14 and 15. The compound resulting from the FUM6 enzyme-catalyzed reaction is proposed to be similar to the FUM8 product but with hydroxyl functions at carbon atoms 14 and 15. Thus, mutant strains of F. verticillioides in which FUM6 has been inactivated are predicted to produce 2-amino-12,16-dimethyleicosane-3-one, i.e. the proposed product of the FUM8 reaction.
Objective
Structural identification of the first two intermediates in the fumonisin biosynthetic pathway by examination of two mutants of F. verticillioides in which FUM cluster genes have been inactivated.
Work plan
Step 1
Comparison of metabolite profiles of F. verticillioides wild-type strain M-3125 with metabolite profiles of FUM6 and FUM8 mutants. Extracts will be screened using LC-MSn in order to point out target intermediates for further structural characterization.
Step 2
The target intermediates will be isolated and purified using combinations of liquid/liquid-extraction and semi-preparative LC. Since the proposed intermediates are similar in structure to the F. avenaceum metabolite 2-AOD-3-ol, the approach for the purification will be based on methodology developed previously at NVI with the necessary adjustments. The structures of the fumonisin biosynthetic intermediates will be determined unequivocally using one- and two-dimensional NMR spectroscopy.

Figure 2. Preparative chromatography of an extract from a dFUM6 mutant strain of Fusarium verticilloides on silica gel