Participants
Section for Chemistry and Toxicology, Norwegian Veterinary Institute, Oslo, Norway (Dr. Silvio Uhlig)
Section for Mycology, Norwegian Veterinary Institute, Oslo, Norway (Dr. Trude Vrålstad, Elin Rolèn)
Background and state of the art
Phylogenetic studies have routinely identified cryptic species within morphological species in various fungal genera including Fusarium and Stachybotrys.What has not been determined in most of these studies is the mechanism or driving force behind the speciation process. In some instances, geographical isolation has been suggested, whereas in other cases these putative cryptic species may be found in the same geographical location and, in some instances, even isolated from the same soil core. According to the literature ‘ecological speciation’ may be defined as the ‘process by which barriers to gene flow evolve between populations as a result of ecologically based divergent selection’.
One group of fungi where ecological aspects may have influenced speciation is within the Claviceps purpurea complex. These are plant-pathogenic fungi that infect the flowers of grasses and cause a disease known as ‘ergot’. The infection results in a replacement of the grass floral tissue with fungal mycelium that grows to form a darkly pigmented survival structure called a sclerotium. Sclerotia of C. purpurea contain potent alkaloids that are toxic to humans and other animals. Previous research conducted at the Academy of Sciences of the Czech Republic on C. purpurea morphology, alkaloid chemistry and genetics identified three distinct groups within the species, describing them as ‘chemoraces.’ Rather than species delimitation based on host range, which was historically common in C. purpurea taxonomy, intraspecific groups were defined based on habitat specialization. The largest group, G1, is associated with land grasses. G2 isolates are associated with grasses in ‘wet and shady’ environments, whereas G3 isolates are found only on grasses in salt marsh habitats. G3 isolates are synonymous with C. purpurea var. spartinae.
It has further been suggested that each genotype produces its specific alkaloid ‘fingerprint’. However, during the last years we have analysed alkaloids in C. purpurea sclerotia from several grasses originating from Southern and Middle Norway. We recognised that the alkaloid profile was highly variable and therfore questioned the connection between alkaloid ‘fingerprint’ and genotype (Figure 1).
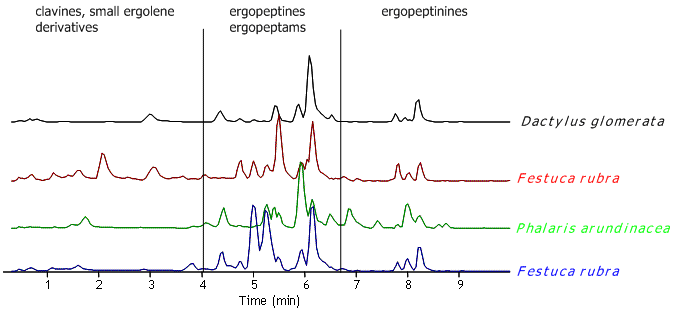
Figure 1. LC-MS chromatograms of extracts from sclerotia picked from grass populations in a habitat in Telemark/South Norway
Objective
Molecular analysis of C. purpurea isolates obtained from sclerotia that were picked from grass populations within confined habitats in Southern Norway in order to determine their genotype, and alkaloid analysis of sclerotia from the same populations.
Work plan
Part 1
Collection of sclerotia from grass populations in confined habitats in Oslo and at Solvik/Telemark, identification of grass species.
Part 2
DNA extraction from fungal isolates obtained from the sclerotia; PCR, sequencing and phylogeny construction based on the ITS- and beta-tubulin genes.
Part 3
LC-MS based analysis of the sclerotia for ergot alkaloids and indole-diterpenoids and comparison of alkaloid fingerprints between genotypes, habitats and grass species.